Using RCy3 with Cytoscape: GSOC Wrap Up
This summer I had the pleasure of working with the National Resource for Network Biology (NRNB as a summer intern as part of Google Summer of Code project. For my project (described more in depth in the previous blog post) I wanted to make workflows for biologists to make their analyses more straightforward by using the programming language R with the Bioconductor package RCy3.
Today I want to discuss my experience with GSOC and working on my project this summer. In a forthcoming blog post I will go into more detail about the workflows that I worked on.
My experience
Overall, working with NRNB as part of Google summer of Code was a great experience. I interacted with many different people and was well supported by my mentor and the overall community. It was a new challenge to work remotely for the whole of a project, but I am happy with how it turned out.
I worked on different aspects of the connection between R and Cytoscape while creating the vignettes as part of this project. I received a lot of help from many different people. Before starting this project I played around with the cyREST API in R and python and Keichiro Ono helpfully answered my questions about getting started with cyREST. One of the workflows that I wanted to work through included a Cytoscape plugin called Enrichment Map and Ruth Isserlin of the Bader Lab (website) provided a lot of troubleshooting and help for this vignette. We selected other Cytoscape applications for use with RCy3 and Scooter Morris was often very helpful in understanding how they were working (and was often an author of these plugins).
The developers of RCy3 were very helpful with this project. Tanja Muetze provided a lot of expertise and encouragement along the way and Paul Shannon and Barry Demchak helped get the project off the ground and helped determine the extent of what would be possible using cyREST with RCy3. Finally, my mentor Augustin Luna was great for discussing the direction of the project, and for providing expertise with web frameworks, R package development and general R coding. The GSOC projects were smoothly coordinated by Alexander Pico, of the NRNB.
The RCy3 package (stands for R to Cytoscape 3 and is actively developed by Tanja Muetze, Georgi Kolishovski, Paul Shannon) uses the CyREST api to allow communication between R and Cytoscape. CyREST now comes with all installations of Cytoscape. It uses the API (application programming interface) from Cytoscape to send and receive information from R. This means that we can send data from R to Cytoscape and also receive information about the graphs that you have made in Cytoscape in R. This is useful for reproducibility, but also if you are analysing networks in ways that are not yet supported by plugins in Cytoscape. )
Working with cyREST
For Cytoscape to be accessible to users of different programming languages an API called CyREST has been developed in Dr. Ideker’s lab. More information about it is available in the paper, in the Github repository and on this website that lists all the commands that are available in the API.
CyREST is now available as part of Cytoscape when you download versions 3.3 and newer. Before that it was a plugin that you could download and install in Cytoscape.
Difficulties with CyREST
Generally I didn’t have many problems using cyREST, but there were a few things that I struggled with:
-
Saving images from Cytoscape via cyREST is dependent on the size of the window that you have open. You can use a parameter “h” which sets the size of the final image, but the width is set automatically. Therefore to get different sized or shaped images it is necessary to manipulate your open Cytoscape window to the shape that you want before running your commands via cyREST.
-
Plugins in Cytoscape that present an API interface do not seem to have a standard verb-action vocabulary. It must be noted that developers that make these plugins are not under any obligation to present an API interface (and it is more work for them). This is not a large difficulty, but it can require some guessing of how plugins can be used via cyREST and what commands are available from a specific plugin. It can also be unclear why certain arguments are available in the Cytoscape graphical interface and not available via cyREST.
Ways to examine commands available via API
Our main goal was to create a reproducible analysis by scripting the use plugins in Cytoscape from R. We used RCy3 to create and display networks, but we also wanted to try accessing plugins used within Cytoscape via R. To do this we identified plugins that present their APIs to cyREST and thus are able to be used on the command line and by the user from R. Unfortunately, the commands available via the API are often not included in the documentation of the plugins or Cytoscape, so to find out what commands are available I needed some ways to investigate what was available. For me, with little experience in javascript, I used three different avenues that I will describe below.
1) Via the command line tool in Cytoscape
Within Cytoscape, commands can be run using the command line dialog.
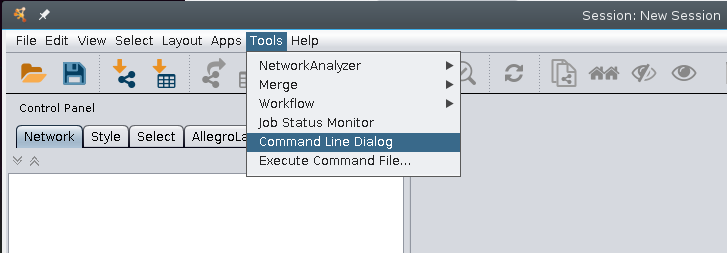
This is also where we can find out about commands by using the help associated with the different plugins and functions of Cytoscape. To examine what information is available, type in help into the command line box and press Enter (or Return).

To look specifically at a specific command you can type help followed by the command (or plugin). If we type help view and press Enter (or Return) here we see:

2) Via your favourite internet browser
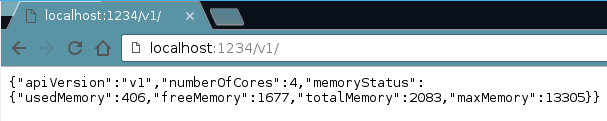
Your browser can also be used to examine the different commands available via the API. The default setting for the cyREST API has Cytoscape at localhost:1234. We will need to direct ourselves to the version 1 of cyREST as well. So if we do that, we get some details about Cytoscape and the API.
This is because cyREST works as a Cytoscape app that exposes the data and functions in Cytoscape via an API. The REST API is an application programming interface that uses HTTP requests to send and receive data, thus it is possible to examine what cyREST is exposing via your web browser.
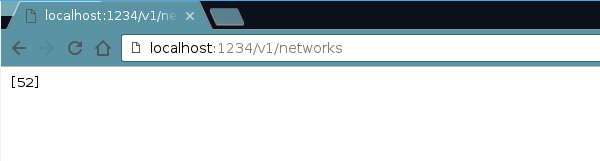
Side note: If we want to see a listing of the networks we have we can type:
Then to examine the possible commands available (like what we did with the command line dialogue above) we can type:

If we want to see the commands available within a set of commands we can type:

You could indeed run commands from here, but this is not very practical and would defeat our purpose. This was just to show how to investigate the plugins and Cytoscape. We will switch to RCy3 in R to run the commands that we want to execute.
3) Using RCy3
In R, I have written code that allows me to check on the commands available and the arguments available for these commands:
library(RCy3)
source("./functions_to_add_to_RCy3/working_with_namespaces.R")
Test out the function to see what commands are available in Cytoscape
cy <- CytoscapeConnection()
getCommandNames(cy)
## [1] "cluster" "clusterviz" "command" "edge"
## [5] "enrichmentmap" "group" "layout" "network"
## [9] "node" "session" "table" "view"
## [13] "vizmap"
(Please note: Your view may be different if you have different plugins installed) See what arguments are available within Cytoscape command “view”
getCommandsWithinNamespace(cy, "view")
## [1] "create" "destroy" "export" "fit content"
## [5] "fit selected" "get current" "list" "set current"
## [9] "update"
Other summer activities: Poster presentation of GSOC work
I presented part of one of the workflows that I was working on at a scientific meeting this summer. The meeting was the Viruses of Microbes meeting in Liverpool, UK. The repository that contains the poster and the code used to generate it are here. This was my first time creating and presenting a poster like this and it was a good challenge. The poster was well received and many jotted down the address for the Github repo.
Second blog post to come with workflows:
Next week I will post second blog post detailing the different workflows that were created. With the help of the RCy3 developers we will be incorporating some of these workflows into RCy3.
Topics within the forthcoming blog post:
- Enrichment Maps using Cytoscape plugin EMap
- Co-occurrence network using microbial data
- Using paxtoolsr to get information from Pathway Commons and display this information in Cytoscape
- Using chemviz to examine the chemical properties and structures of data and creating a network based on chemical similarity
In the meantime please feel free to check out my final work submission which describes the projects and provides links to the individual workflows.